Transcriptome and Metabolome Changes in Hepatic Ischemia-Reperfusion Injury

(A) Hematoxylin-eosin staining of liver tissue samples of the Sham, I1R12, I1R24, and I1R48 groups (magnification, × 200/ × 400; scale bar, 100 mm). (B) Serum concentrations of aspartate aminotransferase (AST) and alanine aminotransferase (ALT). (C) Suzuk
Transcriptomics and metabolomics analysis reveal the characteristic alterations in different stages of hepatic ischemia-reperfusion injury
CHINA, March 3, 2025 /EINPresswire.com/ -- Hepatic ischemia-reperfusion injury (IRI) is a common and unavoidable complication associated with liver transplants that may result in impaired liver function or even post-transplant liver failure. Although various mechanisms have been implicated in the pathogenesis and progression of hepatic IRI, the characteristic alterations occurring at the transcriptome and metabolic levels remain to be investigated.
In this study, published in the Genes and Diseases journal, researchers from Chongqing Medical University employed transcriptomics and metabolomics analyses to identify the differentially expressed genes and metabolites during the early, intermediate, and late phases of hepatic IRI in a mouse model.
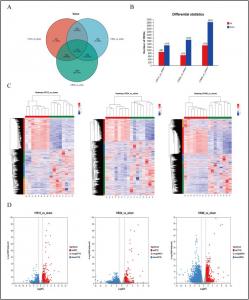
Mouse livers subjected to reperfusion for 12 hrs, 24 hrs, and 48 hrs (corresponding to early, intermediate, and late stages) following 1 hr-ischemia were used for further analysis. Transcriptome data analysis revealed that 1115 differentially expressed genes were co-expressed between the three groups. KEGG and GO analysis revealed the enrichment of glucose and carbohydrate metabolic pathways in early IRI, enrichment of glucose/lipid metabolic pathways and activation of the inflammatory pathway in intermediate IRI, and the enrichment of the lipid metabolic pathways during the late phase of IRI. These results were further validated with Western blotting and qRT-PCR, which revealed the activation of the PI3K/AKT/mTOR pathway.
Metabolomics analysis showed that primary metabolic characteristics of lipid metabolism were altered in the early, intermediate, and late phases of IRI. Furthermore, quantification of free fatty acids in the mouse liver tissues showed a significant decrease in the IR12 and IR24 groups, suggesting a disorder in lipid metabolism.
In conclusion, the key findings of this study show that i) intermediate IRI is characterized by an inflammatory disorder triggered by the overproduction of ROS and the release of DAMPs and pro-inflammatory cytokines, which aggravate apoptosis and hepatocyte damage, ii) the marked upregulation of the PI3K-AKT and HIF-1 pathways in the intermediate phase serves as an adaptive response in regulating anaerobic glycolysis and the inflammatory response to improve IRI, and iii) the glycolysis/gluconeogenesis pathway is altered exclusively during the early phase of IRI. The authors suggest therapeutic interventions targeting glucose and lipid metabolism reprogramming pathways may help mitigate hepatic IRI.
Reference
Title of the original paper: Multi-time point transcriptomics and metabolomics reveal key transcription and metabolic features of hepatic ischemia-reperfusion injury in mice
Journal: Genes & Diseases
Genes & Diseases is a journal for molecular and translational medicine. The journal primarily focuses on publishing investigations on the molecular bases and experimental therapeutics of human diseases. Publication formats include full length research article, review article, short communication, correspondence, perspectives, commentary, views on news, and research watch.
DOI: https://doi.org/10.1016/j.gendis.2024.101465
Funding information:
National Natural Science Foundation of China (No. 82300745)
China Postdoctoral Science Foundation (No. 2023M730442)
Chongqing Postdoctoral Science Foundation of China (No. CSTB2023NSCQ-BHX1003)
Postdoctoral Cultivation Project of the First Affiliated Hospital of Chongqing Medical University (No.CYYY-BSHPYXM-202301)
Chongqing Postdoctoral Innovation Talents Support Program (Chongqing, China) (No. 2309013437264551)
# # # # # #
Genes & Diseases publishes rigorously peer-reviewed and high quality original articles and authoritative reviews that focus on the molecular bases of human diseases. Emphasis is placed on hypothesis-driven, mechanistic studies relevant to pathogenesis and/or experimental therapeutics of human diseases. The journal has worldwide authorship, and a broad scope in basic and translational biomedical research of molecular biology, molecular genetics, and cell biology, including but not limited to cell proliferation and apoptosis, signal transduction, stem cell biology, developmental biology, gene regulation and epigenetics, cancer biology, immunity and infection, neuroscience, disease-specific animal models, gene and cell-based therapies, and regenerative medicine.
Scopus CiteScore: 7.3 | Impact Factor: 6.9
# # # # # #
More information: https://www.keaipublishing.com/en/journals/genes-and-diseases/
Editorial Board: https://www.keaipublishing.com/en/journals/genes-and-diseases/editorial-board/
All issues and articles in press are available online in ScienceDirect (https://www.sciencedirect.com/journal/genes-and-diseases).
Submissions to Genes & Disease may be made using Editorial Manager (https://www.editorialmanager.com/gendis/default.aspx ).
Print ISSN: 2352-4820
eISSN: 2352-3042
CN: 50-1221/R
Contact Us: editor@genesndiseases.com
X (formerly Twitter): @GenesNDiseases (https://x.com/GenesNDiseases)
Genes & Diseases Editorial Office
Genes & Diseases
+86 23 6571 4691
email us here
Visit us on social media:
Facebook
X
LinkedIn
Instagram
YouTube
Other
Distribution channels: Education, Healthcare & Pharmaceuticals Industry, Science
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this article. If you have any complaints or copyright issues related to this article, kindly contact the author above.
Submit your press release